Department
Graduate school of Biomedical science and engineering HANYANG University
Graduate school of Biomedical science and engineering HANYANG University
The birthplace of convergence-type talents
to lead future medicine based on bio-big data
Biomedical informatics is a fusion of the three fields of life science (BT) + medicine + information science (IT).Recently, the amount of data produced in life science experiments has increased tremendously due to the development of new technologies such as biochips and next-generation sequencing analyzers. It has become an era where it is impossible to analyze and interpret the results of experiments and tests without using large-capacity information processing technology or data mining technology developed in information science fields such as computer science and statistics.In particular, the paradigm of future medicine is rapidly changing to personalized medicine based on bio and health care big data as the cost of producing genomic data for an individual is rapidly decreasing. It is very important to nurture future-oriented convergence talents equipped with both the specialized knowledge and technology of It was newly established in 2014 as the Department of Specialty Chemistry designated by the Ministry of Science, ICT and Future Planning.
The educational goal of the Department of Biomedical Informatics is to apply the methodology of Informatics to develop new technologies that enable a deeper understanding of Biology, to nurture specialized knowledge of informatization-based future medicine, and to , through practical education that makes the research results of Bio-Medical Informatics, which converges the three fields of biology, medicine, and informatics, clinically feasible, large-capacity bio-big data (Bio) to be produced in the next-generation medical life field. It aims to nurture convergence specialists essential for the realization of future medicine with information processing technology of Big Data).
1. BT+IT+Medicine Trinity education to nurture convergence-type talents in the three majors of genomic informatics, proteomics, and systems biology
2. essential for hospitals, research institutes, and industries necessary for the next generation of personalized genomic medical care Cultivating practical life medical informatics experts
3. Cultivating global talents who can work in personalized medical teams at overseas university hospitals/research centers/industries

Geneome Informatics
An in-depth study of genotype-phenotype interrelationships such as disease outbreaks, species specificity, and individual differences by precisely analyzing various omics and NGS data such as human, nnanimal and plant genomes and metagenomes
As a result of the Genome Project, which finds the various genetic information possessed by humans and plants and animals, a variety of genomic information including all gene sequences (DNA, mRNA, nsRNA, miRNA...) before protein generation is generated In order to study various life phenomena (differentiation, development, metabolism, drug response, disease mechanisms, etc.) occurring in living things, there is a limit to research at the individual gene level. this came to be Cells, the most basic unit of life, have very large computing power that can independently perceive. Cells gather according to a set time schedule to form tissues, organs, systems, and entities in stages, and various life phenomena must occur precisely, automatically and appropriately. can do. DNA sequence information contains not only sequence information of coding/non-coding genes, but also information on epigenetic modifications, interactions between each protein & DNA, and rapidly changing Nucleoprotein Complex.
With the development of the latest next-generation sequencing (NGS) technology, omics and NGS genome/transcriptome data have become enormously vast, and these genomic data not only affect genetic diseases, but also cancer, chronic diseases, individual diversity and evolution. It turns out that it is a complex working system that gives Various disciplines are being used to analyze and interpret these complex systems, and genome informatics research uses high-performance computers to process, process, and interpret genomic information using informatics methods such as statistics and computer science to derive useful information. As a research field, students acquire informational knowledge and skills such as computer programming and data mining based on their knowledge of biology and medicine.
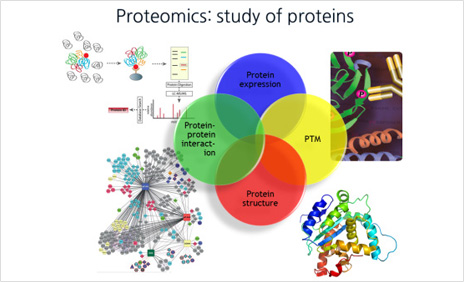
Proteome Informatics
Research on protein function and disease mechanisms in vivo based on an understanding of proteomic expression, quantification, interaction, and modification through proteomic analysis and data analysis and interpretation
In the past, proteins in cells were studied one at a time, but with the development of innovative technology, it is now possible to study all proteins in cells at once. More specifically, identification of proteins to find out which proteins are expressed in cells, quantification to determine how the amount of each protein changes in the disease state compared to the normal state, and protein function It refers to the analysis of three-dimensional structures, which is essential for determining, post-translational modification analysis that studies the modification of proteins in cells after protein expression, and analysis of protein-protein interactions.
In particular, it is the development of mass spectrometry technology that has provided a breakthrough in proteomic analysis, and is currently being used extensively in all stages of analysis, such as proteomic identification, quantification, structure, interaction, and PTM. In particular, mass spectrometry is the only technology capable of rapidly performing PTM analysis in large quantities. However, since various databases and software must be used to interpret the mass spectrometry results, an in-depth understanding of these is essential for research. In addition, as the data production speed increases in recent proteomic research, the use of computing technology for processing large amounts of data is also becoming an important issue.
Meanwhile, the introduction of proteogenomics research is being attempted in connection with the development of next-generation sequencing (NGS) technology for genome and transcriptome research. It is intended to contribute to uncovering the cause of disease by deriving new information that is not yet available. For such a study, not only requires an integrated knowledge to integrate genome, transcriptome, and proteomic analysis results, but also understands distributed computing technology that can effectively process NGS and proteomic analysis because each of NGS and proteomic analysis produces large amounts of data. is required
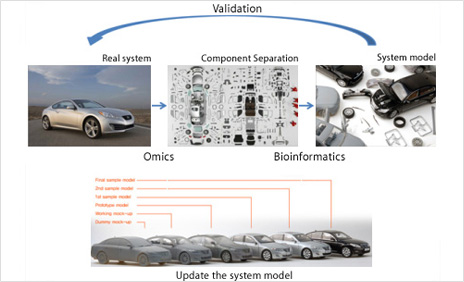
Systems Biology
Research on understanding complex disease mechanisms and finding biomarkers by integrating multi-omics information such as the produced genome/transcriptome/proteome and analyzing the interaction relationship between genes, proteins, and the environment
Systems biology is a field of study that seeks to understand life phenomena within complex systems. Previous studies in the field of general biology have focused on one or two molecules of interest using a simplified model system. As a result, it is true that there were many theories and methods that worked well in vitro, but did not apply to the living body. This is a problem caused by trying to understand living things too simply, while ignoring complex systems. In order for a person to live, he has to have relationships with many other people around him. Because there is that person in the midst of living with many people, such as family, friends, co-workers, etc., ignoring all these relationships and analyzing only one person may make it impossible to understand or misunderstand the person itself. The approach to life is the same. One protein is said to be correlated with at least six other proteins. Therefore, in order to accurately understand the function of a specific protein or gene in vivo, it is necessary to analyze the system as it is, instead of reducing and simplifying the system. That is the direction systems biology is ursuing.
The figure above is a schematic representation of systems biology. In order to understand the real world with our current technology, there is no other way than to tear apart each component of the real world. Because in many cases we do not know what a component does (there are only about 8,000 genes for which we know its function), it is usually possible to extract all components from normal and disease, control and treatment groups, and compare them. Check the components. Changes in expression of genes were analyzed by microarray technique, etc., proteins were analyzed using 2DE and mass spectrometry, and differences in nucleotide sequences were analyzed using NGS, etc. Now let's weave these data more systematically. By making a new model through each component rather than a simple comparison, verifying the model by comparing the model with the real world, and updating the model continuously, it is possible to make a model closer to the real world, and thus the understanding of life phenomena is also improved. because it will be Therefore, it can be said that systems biology is a field that requires a variety of knowledge not only for understanding various omics data, but also for interaction with each molecule and a mathematical model using the same.